Tumor
癌症序列变异解释和报告的标准和指南(ASCO和CAP联合推荐)
肺癌分析插件Can_28修复问题
Chom化疗插件及bug修复
Brca插件
FACTERA-fusionGene
SEGF-挖掘NGS中融合基因的新方法
maftools-肿瘤突变数据可视化神器
从数据库中获取免疫相关基因(IRGs)
数据库10KIP—基于ImmPort免疫组学数据挖掘
TCGA数据分析
下载分析TCGA数据库的数据
差异表达分析(limma & edgeR & DESeq2)
TCGA数据的规律【更新中】
生存分析
生存模型构建
突变数据
本文档使用 MrDoc 发布
-
+
up
down
首页
Chom化疗插件及bug修复
目前肿瘤多个实验策略分布: - (化疗、乳腺癌2基因、肺癌Can28、个体化用药50基因Chp2、中创100位点均是采用多重PCR进行扩增测序;AIO为捕获测序) - (化疗、乳腺癌2基因、中创100位点均为germline,仅肺癌Can28、个体化用药50基因Chp2以及AIO panel为体细胞突变) 化疗插件,是用TVC检测的变异。 1、分析流程 (1)通过脚本bin/get_chmo_bam.pl遍历basecaller_results目录下的bam文件,判断bam文件中的SM信息中是否包含chmo(不区分大小写),并链接到rawdata目录中; (2)运行主程序 ``` ## perl ${PLUGIN_PATH}/chmo_pipeline.pl ${ANALYSIS_DIR} ${RESULTS_DIR} ${reference} ${PLUGIN_PATH}/data/amplican.bed ${PLUGIN_PATH}/data/target.bed perl /results/plugins/Chom/chmo_pipeline.pl /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271 /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629 /results/referenceLibrary/tmap-f3/hg19/hg19.fasta /results/plugins/Chom/data/amplican.bed /results/plugins/Chom/data/target.bed ``` 生成total.sh,包括每个样本的shell命令。如:sh /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167.sh; - 扩增子文件amplican.bed(共13个): ``` chr1 20915650 20915789 Came_01 chr1 20931409 20931528 Came_02 chr1 97915564 97915681 Came_03 chr1 11856334 11856473 Came_04 chr2 234668802 234668928 Came_05 chr7 87138600 87138720 Came_06 chr7 87160520 87160659 Came_07 chr11 67352640 67352771 Came_08 chr15 51502793 51502926 Came_09 chr19 44055682 44055818 Came_10 chr19 44057511 44057633 Came_11 chr19 45923609 45923742 Came_12 chr22 42526642 42526766 Came_13 ``` 对应的目标捕获区间文件target.bed文件 ``` chr1 20915671 20915766 Came_01 chr1 20931432 20931505 Came_02 chr1 97915587 97915658 Came_03 chr1 11856357 11856450 Came_04 chr2 234668825 234668908 Came_05 chr7 87138623 87138697 Came_06 chr7 87160543 87160638 Came_07 chr11 67352661 67352748 Came_08 chr15 51502816 51502903 Came_09 chr19 44055703 44055797 Came_10 chr19 44057534 44057611 Came_11 chr19 45923631 45923719 Came_12 chr22 42526662 42526746 Came_13 ``` (3)每个样本的shell具体过程 ``` # step1, easy_trim.pl对下机的bam文件进行修剪。筛选出序列长度高于50的reads,对其末端20bp进行修剪,并输出到Tag167.clean.bam文件中,并统计clean_rate到Tag167/Tag167.clean.stat中。 perl /results/plugins/Chom/bin/easy_trim.pl /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/rawdata/Tag167_rawlib.basecaller.bam /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167.clean > /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167.clean.stat # step2,比对,sort /results/plugins/Chom/tools/tmap mapall -n 12 -g 3 -a 0 -i bam -o 2 -f /results/referenceLibrary/tmap-f3/hg19/hg19.fasta -r /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167.clean.bam -v -Y -u --prefix-exclude 5 stage1 map4 > /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167.mapped.bam && rm /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167.clean.bam /results/plugins/Chom/tools/samtools sort /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167.mapped.bam /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167.mapped.sorted && rm /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167.mapped.bam # step3, 统计Q17和Q20并过滤。 判断read的Q20比例超过该read长度的50%,则保留,并用于统计过滤后的Q17和Q20比例。如Q20比例低于60%,给出WARNING; perl /results/plugins/Chom/bin/get_sm_q20.pl /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167.mapped.sorted.bam /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629 Tag167 > /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167.q20.stat # step4, 过滤reads长度低于30的,并统计target区覆盖度,ontarget率等等。生成Tag167.fixed.bam,建index,统计各位点depth,以及扩增子平均深度。对于ontarget率低于0.7的在Tag167.stat文件末尾给出WARNING。 perl /results/plugins/Chom/bin/bam_qc.pl /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167.qual.bam Tag167 /results/plugins/Chom/data/amplican.bed /results/plugins/Chom/data/target.bed /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167 /results/plugins/Chom/tools/samtools index /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167.fixed.bam perl /results/plugins/Chom/bin/bam_depth2.pl /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167.fixed.bam > /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167.stb.depth perl /results/plugins/Chom/bin/amp_depth.pl /results/plugins/Chom/data/target.bed /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167.stb.depth > /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167.stb # step5, TVC变异检测; python /results/plugins/Chom/tools/variantCaller/variant_caller_pipeline.py -i /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167.fixed.bam -o /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167variant -B /results/plugins/Chom/tools/variantCaller -n 10 -r /results/referenceLibrary/tmap-f3/hg19/hg19.fasta -b /results/plugins/Chom/data/target.bed -p /results/plugins/Chom/data/targetseq_germline_lowstringency_p1_parameters.json -s /results/plugins/Chom/data/100.hotspot.vcf # step6, split多等位基因位点并校正TVC检测的位点(注意:最后参数指定的mppana.txt在该步骤未用上的); perl /results/plugins/Chom/bin/splitVcfMuliAlt.pl /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167variant/TSVC_variants.vcf /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167variant/TSVC_variants.split.vcf /results/plugins/Chom/data/100.hotspot.vcf /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167variant/Tag167.mppana.txt # step7, mpileup分析; /results/plugins/Chom/tools/samtools mpileup -BQ0 -d 100000 -f /results/referenceLibrary/tmap-f3/hg19/hg19.fasta -l /results/plugins/Chom/data/target.bed /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167.fixed.bam > /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167variant/Tag167.pileup perl /results/plugins/Chom/bin/mpileupAnalysis.pl /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167variant/Tag167.pileup /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167variant/Tag167.mppana.txt # step8, 提取变异信息 #perl /results/plugins/Chom/bin/extractInfoTvcVcf.pl /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167variant/TSVC_variants.split.vcf /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167variant/Tag167.allvar.txt perl $Bin/bin/extractInfoTvcVcf_new.pl $out_dir/$sample_name/$variant/TSVC_variants.split.vcf $out_dir/$sample_name/$variant/$sample_name.mppana.txt $out_dir/$sample_name/$variant/$sample_name.allvar.txt rm /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/*.sam # step9, 关联到用药字典dictionary.txt(同时对chr7:87160618:ABCB1:c.2677T>A/G和chr2:234668879:UGT1A1:c.-52TA[7]做处理)。该步对这两个位点修改bug,用程序chemo_new.pl。 #perl /results/plugins/Chom/bin/chemo.pl /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167variant/Tag167.allvar.txt /results/plugins/Chom/data/dictionary.txt /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167variant perl /results/plugins/Chom/bin/chemo_new.pl /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167variant/Tag167.allvar.txt /results/plugins/Chom/data/dictionary.txt /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167variant # step10,结果上传到报告系统 perl /results/plugins/Chom/bin/uploadResult.pl /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167variant/chemo_simple.txt /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167variant/chemo_output.txt CHMO-LCM-20180920-167 /results/analysis/output/Home/Auto_user_2456177-5-0049-125-P5-HiQ-pooling-P5-Can28-BRCA-CHMO-20180920_264_271/plugin_out/Chom_out.629/Tag167/Tag167variant ``` 2、bug1: 完善过滤条件:耳聋100位点也是多重PCR测序的,判断了是否每个hotspot均在TSVC_variants.vcf文件中是否存在,如果不存在,判断该位点的深度是否>=30X,如果为真,则基于频率进行判断。 (1)下面为hotspot文件,/results/plugins/Chom/data/100.hotspot.vcf ``` ##fileformat=VCFv4.1 ##allowBlockSubstitutions=true #CHROM POS ID REF ALT QUAL FILTER INFO chr1 11856378 . G A . . OID=MTHFR:c.665C>T;OPOS=11856378;OREF=G;OALT=A;OMAPALT=A chr1 20915701 . A C . . OID=CDA:c.79A>C;OPOS=20915701;OREF=A;OALT=C;OMAPALT=C chr1 20931474 . G A . . OID=CDA:c.208G>A;OPOS=20931474;OREF=G;OALT=A;OMAPALT=A chr1 97915614 . C T . . OID=DPYD:c.1905+1G>A;OPOS=97915614;OREF=C;OALT=T;OMAPALT=T chr2 234668879 . C CAT . . OID=UGT1A1:c.-52TA[7];OPOS=234668879;OREF=C;OALT=CAT;OMAPALT=CAT chr2 234668880 . ATATATATATAT ATATATATATATAT,ATATATATATATATAT . . OID=UGT1A1:c.-52TA[7],UGT1A1:c.-52TA[8];OPOS=234668881,234668881;OREF=TATATATATATA,TATATATATATA;OALT=TATATATATATATA,TATATATATATATATA;OMAPALT=ATATATATATATAT,ATATATATATATATAT chr7 87138645 . A G . . OID=ABCB1:c.3435T>C;OPOS=87138645;OREF=A;OALT=G;OMAPALT=G chr7 87160618 . A C,T . . OID=ABCB1:c.2677T>G,ABCB1:c.2677T>A;OPOS=87160618,87160618;OREF=A,A;OALT=C,T;OMAPALT=C,T chr11 67352689 . A G . . OID=GSTP1:c.313A>G;OPOS=67352689;OREF=A;OALT=G;OMAPALT=G chr15 51502844 . A C . . OID=CYP19A1:c.*161T>G;OPOS=51502844;OREF=A;OALT=C;OMAPALT=C chr19 44055726 . T C . . OID=XRCC1:c.1196A>G;OPOS=44055726;OREF=T;OALT=C;OMAPALT=C chr19 44057574 . G A . . OID=XRCC1:c.580C>T;OPOS=44057574;OREF=G;OALT=A;OMAPALT=A chr19 45923653 . A G . . OID=ERCC1:c.354T>C;OPOS=45923653;OREF=A;OALT=G;OMAPALT=G chr22 42526694 . G A . . OID=CYP2D6:c.100C>T;OPOS=42526694;OREF=G;OALT=A;OMAPALT=A ``` (2)hotspot文件共有14个行,共13位点,经过对433个样本的TSVC_variants.vcf文件检查, ``` ls Auto*/result/*/result/Tag*/*variant/TSVC_variants.vcf|wc -l #总样本数 for i in `cat /results/plugins/Chom/data/100.hotspot.vcf|grep -v '^#' |awk '{split($8,x,/;/);split(x[1],y,/=/);print y[2]}'`;do echo -e $i" \c";grep $i Auto*/result/Tag*/*variant/TSVC_variants.vcf|wc -l;done #每个位点样本覆盖情况 grep 'UGT1A1:c.-52TA' Auto*/result/*/result/Tag*/*variant/TSVC_variants.vcf|wc -l #'UGT1A1:c.-52TA'位点按该语句统计; ``` 其中UGT1A1:c.-52TA有324个,XRCC1:c.1196A>G只有117个,ERCC1:c.354T>C有432个,CYP2D6:c.100C>T共有357个;其余位点均有433个。 修复:不过在TSVC_variants.split.vcf文件中已经修复(即脚本/results/plugins/Chom/bin/splitVcfMuliAlt.pl)。 bug2: 化疗结果发现一个现象:rs2032582这一个位点,二代结果给出AT、AC两个结果,sanger结果为CT;查后台结果该突变检测无误;所以我们考虑应该是这个点在基因型判断的法则上写的有问题,导致显示到前端的结果异常。请抽空帮忙修正下该异常,最好这周可以修正(一方面报给患者一直错不好,一方面马上转检验),可以跟建文讨论 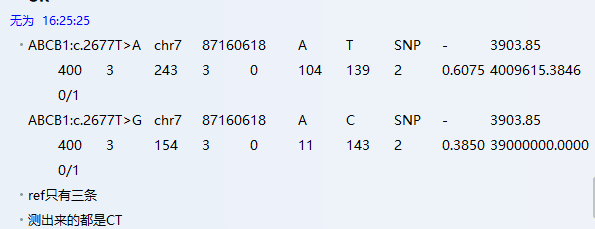  该位点错误示例样本:CHMO-YXS-20180831-140,在RunID: /results/analysis/output/Home/Auto_sn247560054_sn247560054-855-P30-HiQ-Pooling-Can28_CHMO_BRCA-PGX-180831_236_179/plugin_out/Chom_out.484/Tag140/Tag140variant 修复:经过对13个样本一代验证,其中12个二代同时检出该位点为AC、AT两种杂合,一代验证结果为CT,对应的mpileup分析结果中频率也是C和T高(CT之和超过0.98,且ref上A的频率低于0.02),另一个样本二代检出为AC,无AT突变,一代验证结果和二代一致也为AC杂合突变。对这个位点修改bug,用程序chemo_new.pl。是在原程序chemo.pl中加入对chr7:87160618-AC和chr7:87160618-AT的判断,如果都存在,则修改为chr7:87160618-CT输出结果,而不再是AC和AT均输出。 bug2: 化疗这边还有一个紧急问题需要你帮忙测试解决下:UGT1A1,c.(TA)6>(TA)5/(TA)7/(TA)8,rs8175347,这一个点会存在部分样本检测不准,如图: 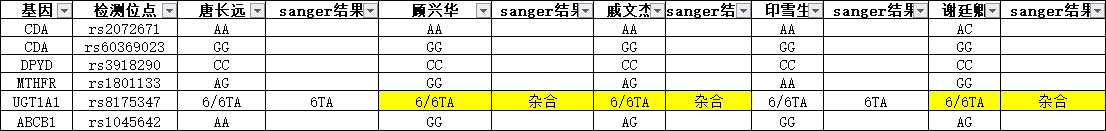  在RunID: /results/analysis/output/Home/Auto_sn247560054_sn247560054-855-P30-HiQ-Pooling-Can28_CHMO_BRCA-PGX-180831_236_179/plugin_out/Chom_out.484/Tag138/Tag138variant sanger纯合和杂合图 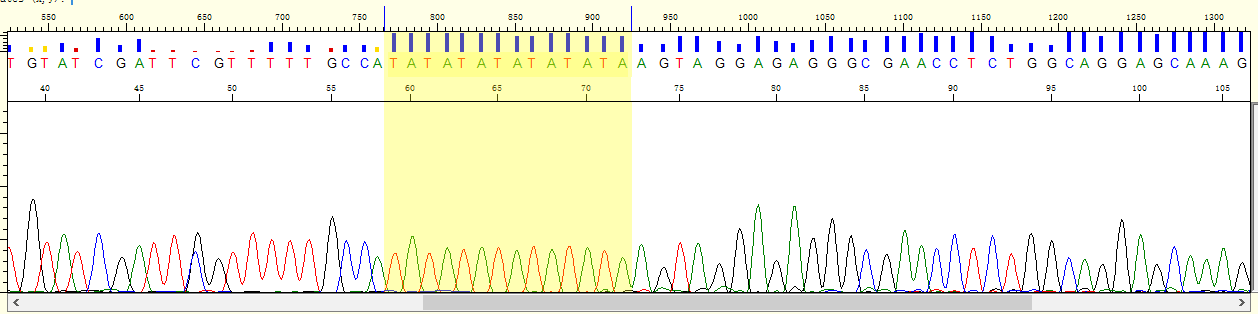  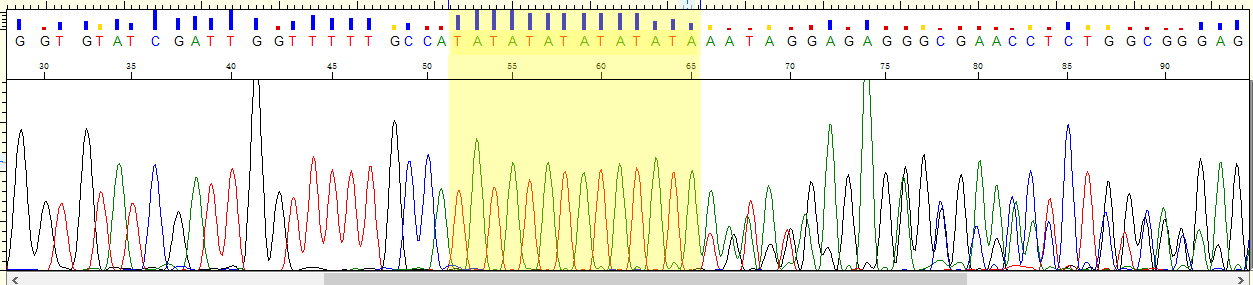 这个问题,药物那边也存在,目前是通过重新设定基因型判断阈值来解决的,参考下按药物基因组的阈值设定,然后重分析这5个样本,看看这个重分析结果和一代结果一致性如何; 药物那边的处理方式是: 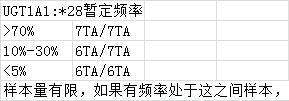 最佳解决方案: 统计好所有化疗检测的该位点(也可以加入药物那边检测的该位点),1、绘制不同基因型的频率分布图(直接设置阈值);2、应用贝叶斯理论(同耳聋位点检测)进行基因型判定; 要用第二种方法,就需要先收集数据,汇总以前所有测的CHOM的数据,由于阳性一代验证的较少,安排阳性样本的一代验证。覆盖从低到高的频率(来自mppana.txt文件) **以下为第二种方法校正**:结果位于文件:Chom_UGT1A1-20180919.xlsx中。通过对86个样品的验证1代结果,并依据贝叶斯定理获得0-1频率范围内的各基因型的可能性。  加入脚本到流程中:chemo_new.pl和extractInfoTvcVcf_new.pl,分别替换原脚本chemo.pl和extractInfoTvcVcf.pl。 运行方式:(extractInfoTvcVcf_new.pl多加了一个mpileup的解析结果作为第二个参数) ``` perl $Bin/bin/extractInfoTvcVcf_new.pl $out_dir/$sample_name/$variant/TSVC_variants.split.vcf $out_dir/$sample_name/$variant/$sample_name.mppana.txt $out_dir/$sample_name/$variant/$sample_name.allvar.txt perl $Bin/bin/chemo_new.pl $out_dir/$sample_name/$variant/$sample_name.allvar.txt $Bin/data/dictionary.txt $out_dir/$sample_name/ $variant ``` 在第一个脚本中,对TSVC_variants.split.vcf变异结果文件中的位点chr2:234668879为C>CAT时,进行频率和基因型的校正。 |af | gt| | --- | --- | |af<=0.05 | 0/0| |0.1<af<=0.65 | 0/1| |af>=0.9 | 1/1| |others | unknow/sanger|
laihui126
2023年1月9日 14:24
分享文档
收藏文档
上一篇
下一篇
微信扫一扫
复制链接
手机扫一扫进行分享
复制链接
关于 MrDoc
觅道文档MrDoc
是
州的先生
开发并开源的在线文档系统,其适合作为个人和小型团队的云笔记、文档和知识库管理工具。
如果觅道文档给你或你的团队带来了帮助,欢迎对作者进行一些打赏捐助,这将有力支持作者持续投入精力更新和维护觅道文档,感谢你的捐助!
>>>捐助鸣谢列表
微信
支付宝
QQ
PayPal
下载Markdown文件
分享
链接
类型
密码
更新密码